We have mapped the protein interaction networks comprising the target of rapamycin (TOR) and SNF1-related (SnRK1) kinases with the aim to understand how these networks regulate plant growth and stress resilience in relation to carbon and nitrogen availability.
Although the TOR pathway is conserved across eukaryotes, plants developed unique adaptations to this pathway to cope with their autotrophic and sessile nature. We generated for the first time a comprehensive TOR signaling network in plants, elucidating both evolutionarily conserved as well as novel plant-specific links, covering a broad range of biological processes such as protein and nucleotide biosynthesis, autophagy, auxin signaling, chloroplast development, lipid metabolism, and senescence.
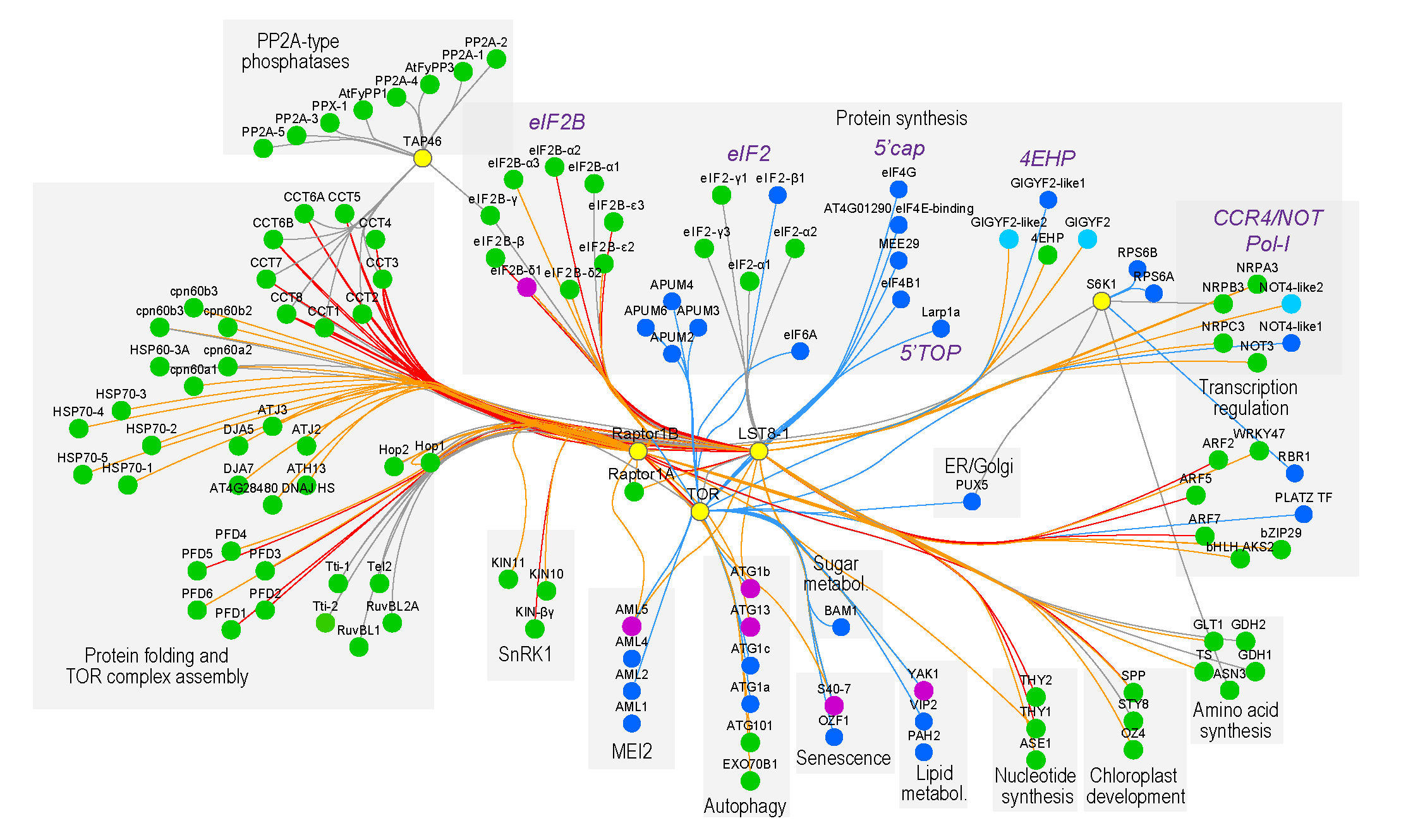
During energy-depleting stress conditions, SnRK1 stimulates catabolic reactions while repressing energy-consuming anabolic processes, shifting the balance from growth to survival. We also mapped SnRK1 regulatory networks, both under different carbon or nitrogen conditions, and this by combining AP-MS, proximity labeling, PTM and cross linking MS analyses.
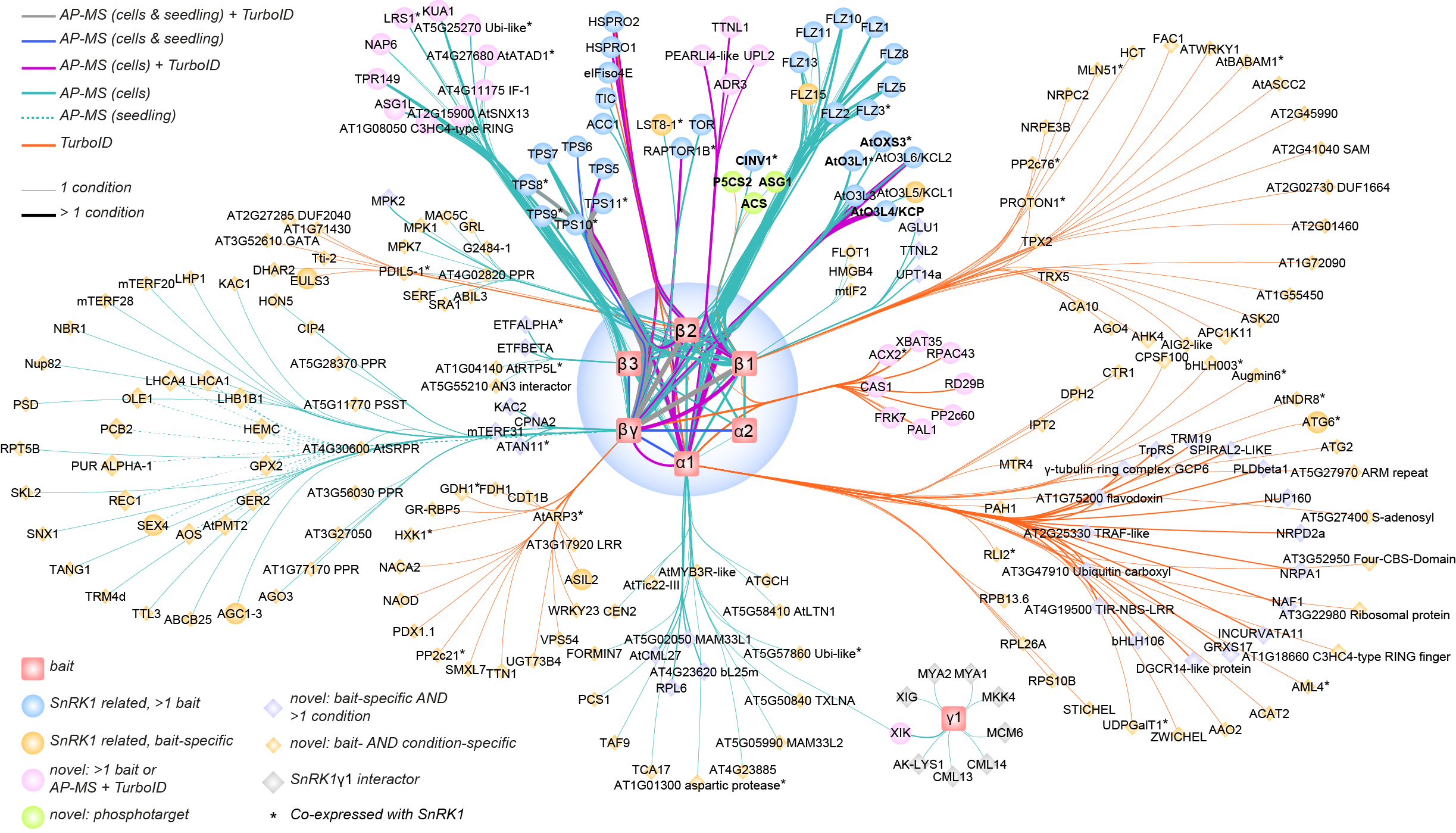
Our networks are rich resources for hypothesis-driven research. Currently, we are studying the function of upstream regulators and downstream targets of TOR and SnRK1 activity we recently isolated. On the other hand, we are translating our network data into beneficial phenotypes. We engineer Arabidopsis plants with the aim to enhance growth, stress resilience or nitrogen use efficiency. Finally, we transfer promising genotypes into crops.