Our interactomics tools are applied to discover new players in the TOR/SnRK1 regulatory pathway that links energy and nutrient (C/N) status with plant growth.
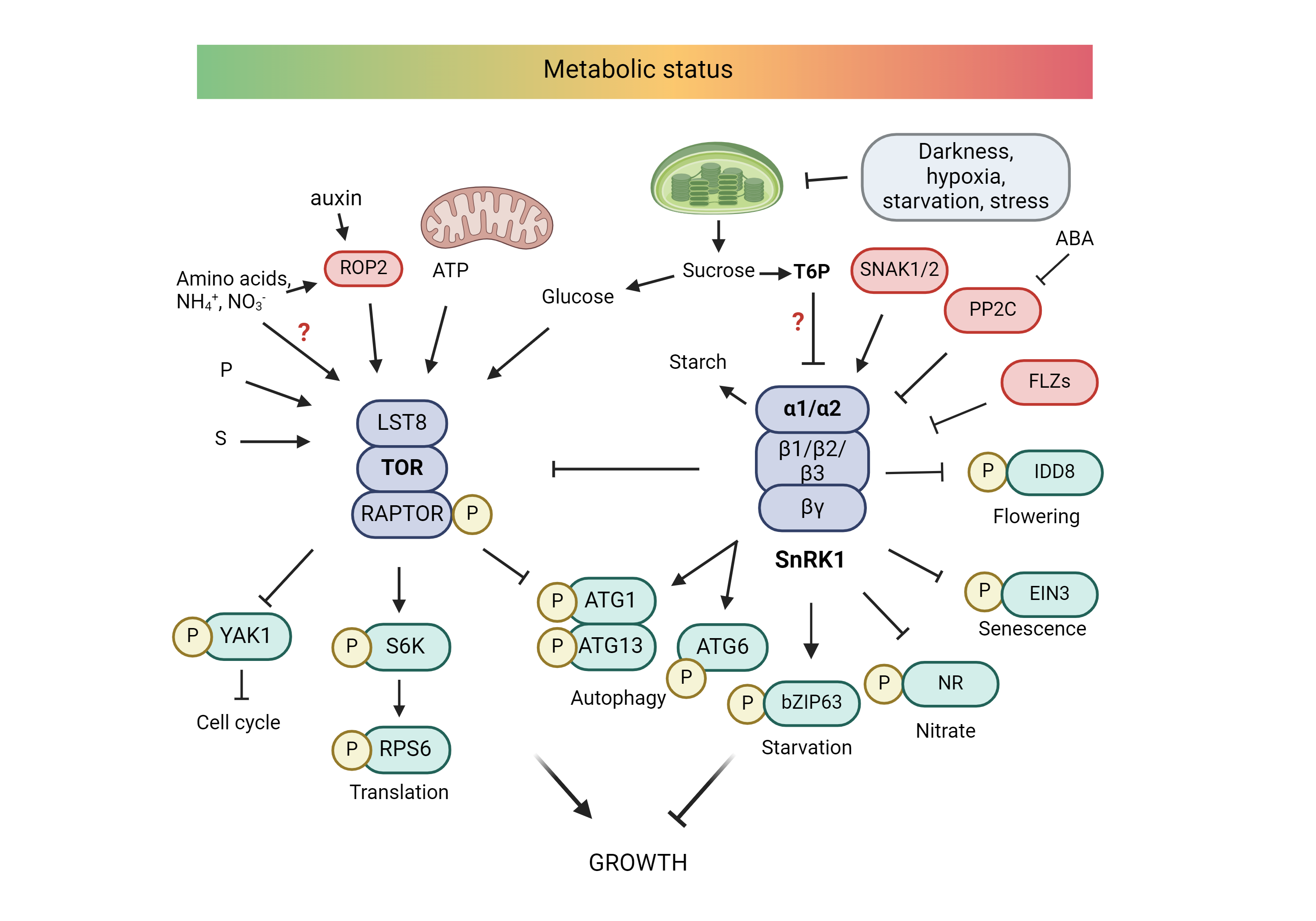
The TOR/SnRK1 pathway has been shown to be involved in yield and stress tolerance in plants. The TOR kinase is a central regulatory hub that translates environmental and nutritional information into permissive or restrictive growth decisions. The central metabolic regulator SnRK1 controls plant growth and survival upon activation by energy and nutrient depletion, while also gatekeeping developmental transitions and nutrient allocation between source and sink tissues. Despite significant progress made in recent years, many questions still remain on how TOR/SnRK1 senses cellular energy and nutrient levels in plants, and how it transduces this information to ensure optimal growth and survival. Therefore, we are mapping proteome-wide TOR/SnRK1 signaling networks in Arabidopsis in relation to nutrient (C/N) availability. We combined phosphoproteomics, affinity purification, proximity labeling and cross linking mass spectrometry to shed light on the interaction landscape, structure and post-translational modification state of the core heterotrimeric TOR and SnRK1 kinase complexes. At the intersection of this targeted interactomics approach, we discovered a strong association of SnRK1 with class II T6P synthase-like proteins. Biochemical and cellular assays showed that these proteins function as negative regulators of SnRK1, and might have a role as carbon sensors that integrate T6P levels with SnRK1 activity.
In parallel we follow a similar approach to explore if SnRK1 also senses nitrogen starvation in the cell, and here we could discover a connection with the endoplasmic reticulum. Increasing nitrogen fertilizer applications and the selection of high-yield crops have sustained our growing world population. However, the future challenge is to restrict fertilizer use, while maintaining crop yield to prevent further land clearing for farming. To facilitate crop growth with less N, we want to upregulate nitrogen utilization efficiency (NUtE) in plants through a better understanding of the TOR/SnRK1 crosstalk in response to the nitrogen status. In addition to the downstream targets, we study how the TOR and SnRK1 kinases are upstream regulated in response to the nitrogen state. Our network data are translated into Arabidopsis to screen for genotypes with better nitrogen efficiency. Finally, we try to translate our findings into crops.
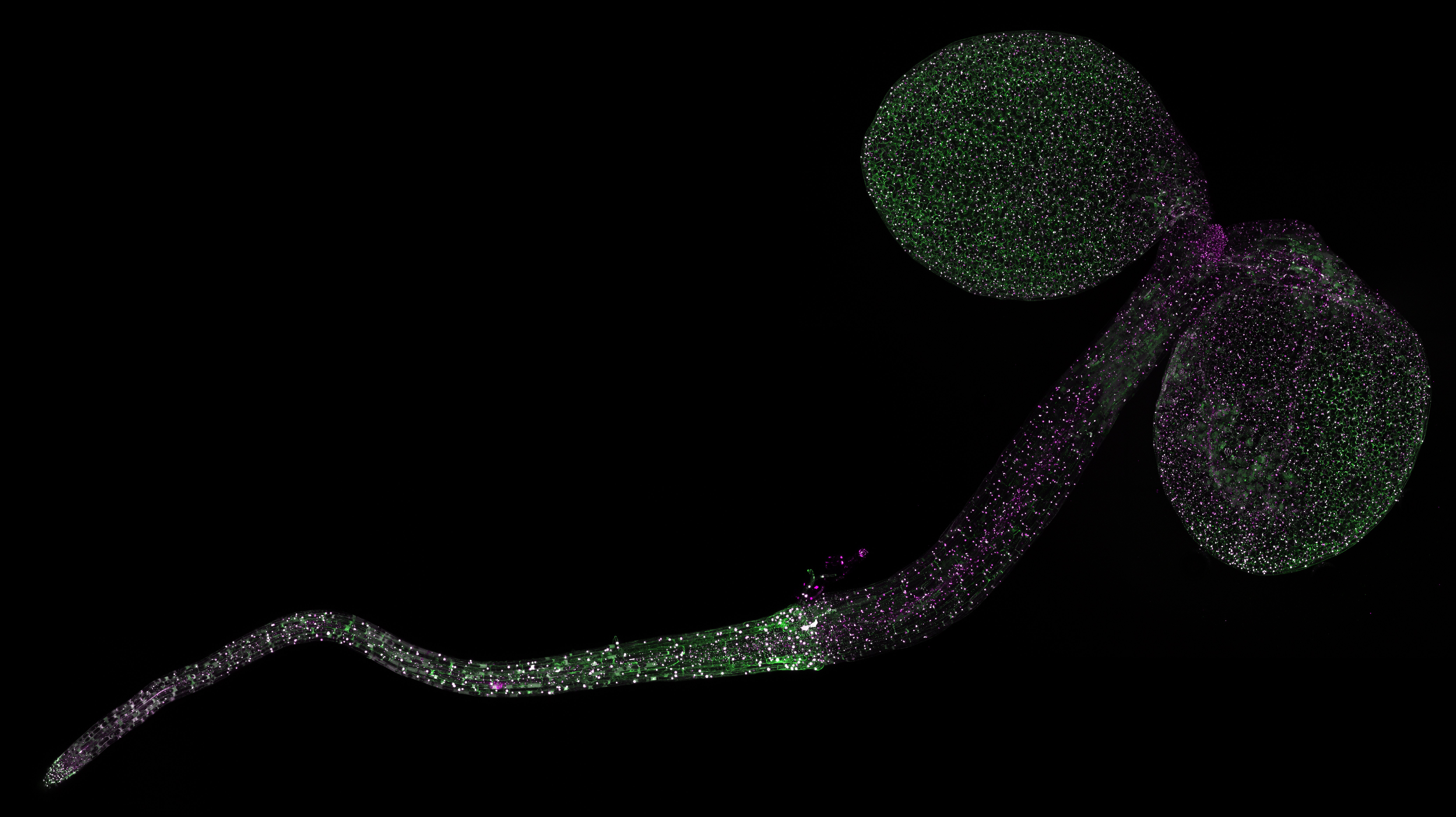
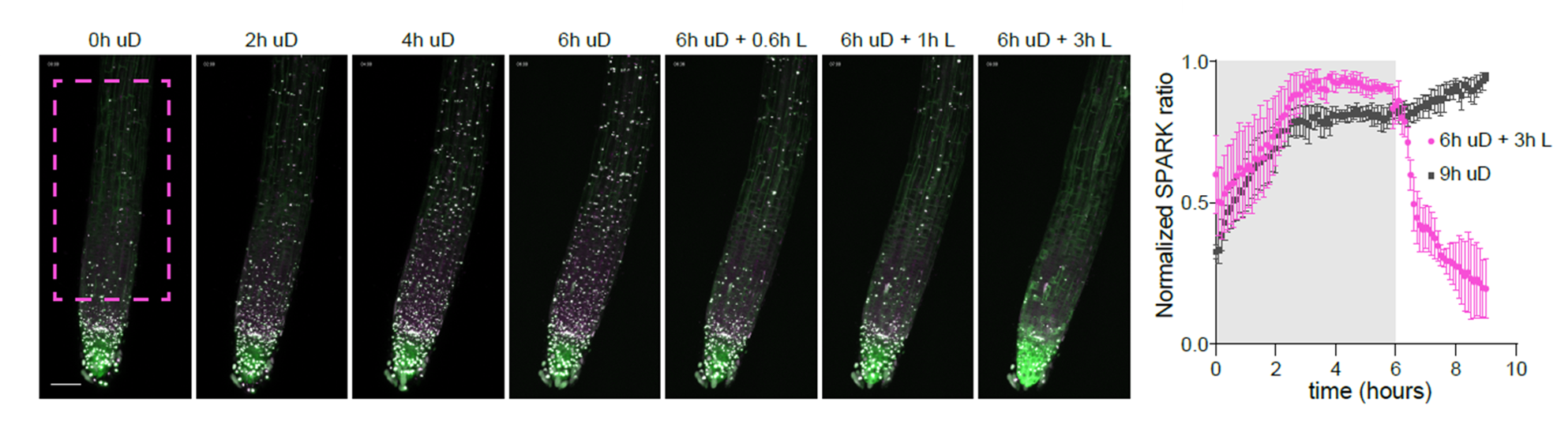
For in planta monitoring of SnRK1 activity, we developed a reversible SPARK-based in vivo SnRK1 kinase reporter. Upon SnRK1-dependent phosphorylation, aggregation of fluorescent droplets is induced, shedding light on SnRK1 kinase activity at tissue level (Safi and Smagghe et al, Plant Cell, 2023).